An international research consortium, led by Fujian Agriculture and Forestry University and the Beijing Genomics Institute has completed the first genome sequence of the diamondback moth, the most destructive pest of Brassica crops.
The genome sequence was completed as part of a study that was published online on Sunday, in Nature Genetics, and may provide new ways to develop sustainable pest management.
The diamondback moth, also known as Plutella xylostella, feeds on important food crops such as rapeseed, cauliflower and cabbage. It has developed resistance against more than 50 insecticides, including DDT and Bt toxins, making the use of chemicals ineffective. It is estimated that the total cost for damage and management associated with this pest is $4-5 billion per year worldwide.
"The completed genome sequencing of diamondback moths will lay a solid foundation for tracking the evolutionary mechanisms of how an insect evolves to develop resistance against many insecticides." said Professor You Minsheng, vice president of Fujian Agriculture and Forestry University and head of the research team. "The work here also provides an invaluable resource for scientists to better understand the reasons the diamondback moth is such a serious pest and how new strategies can be developed to control insect pests."
Professor Wang Jun, executive director of Beijing Genomics Institute, said: "The availability of a reference genome for a species is extremely important in the deeper understanding of its biology and evolution. We are pleased to be part of this consortium and have the first publicly accessible database of the diamondback moth genome. I expect we could translate our achievements into real action for sustainable pest management in the near future".

 Li Na on Time cover, makes influential 100 list
Li Na on Time cover, makes influential 100 list
 FBI releases photos of 2 Boston bombings suspects
FBI releases photos of 2 Boston bombings suspects
 World's wackiest hairstyles
World's wackiest hairstyles
 Sandstorms strike Northwest China
Sandstorms strike Northwest China
 Never-seen photos of Madonna on display
Never-seen photos of Madonna on display
 H7N9 outbreak linked to waterfowl migration
H7N9 outbreak linked to waterfowl migration
 Dozens feared dead in Texas plant blast
Dozens feared dead in Texas plant blast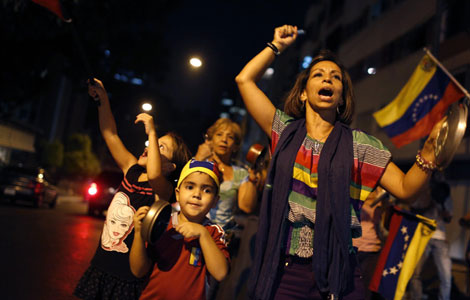
 Venezuelan court rules out manual votes counting
Venezuelan court rules out manual votes counting